[Deep Tech] DNA Search, From Lists to Graphs
Input
Modified
DNA search engine “MetaGraph,” shifting from list-based retrieval to graph-based matching A structural breakthrough improving speed, accuracy, and cost simultaneously, expanding from life sciences into educational search Learning data likewise requires path-based exploration and systematic data governance
This article is a reconstruction tailored to the Korean market based on a contribution to the SIAI Business Review series published by the Swiss Artificial Intelligence Institute (SIAI). The series aims to present researchers’ perspectives on the latest issues in technology, economics, and policy in a manner accessible to general readers. The views expressed herein are those of the author and do not necessarily reflect the official position of SIAI or its affiliated institutions.
In 2025, life science research reached a turning point that fundamentally reshaped the way data is searched. At the center of this shift is MetaGraph, a DNA search engine that integrates 18.8 million DNA and RNA sequences into a single interconnected network. MetaGraph does not simply list information. It searches sequences by tracing “paths” formed by linking k-mers, fragments of nucleotides, placing emphasis on “how things are connected” rather than merely “what exists.” This shift represents more than a technological upgrade; it marks a transformation in how data structures themselves are interpreted. By reducing redundancy across massive sequence datasets and tracing connected paths, MetaGraph enhances accuracy while scaling efficiently.

A New Architecture Enabled by DNA Search Engines
Traditional list-based search systems merely enumerated results. MetaGraph, by contrast, aggregates thousands of short sequences and integrates weak signals into coherent patterns. What determines the result is not an individual token but the order and relationships among sequences. Lists sever the flow of data, while graphs reveal relational structure.
The advantages of this architecture become increasingly pronounced as data volumes grow. As the biological database GenBank expands rapidly year after year, MetaGraph enables comprehensive analysis of vast sequences ranging from viruses to humans in a single search. Instead of loading individual data files, the system simultaneously retrieves related information through connected paths. At the core of MetaGraph lies the de Bruijn graph. Sequences are divided into fixed-length fragments and connected through overlapping regions to form paths. Identical fragments appearing across multiple datasets are stored once and annotated by color to indicate provenance, thereby minimizing redundancy. This compressed structure allows fast and accurate searches even across petabase-scale sequence data.
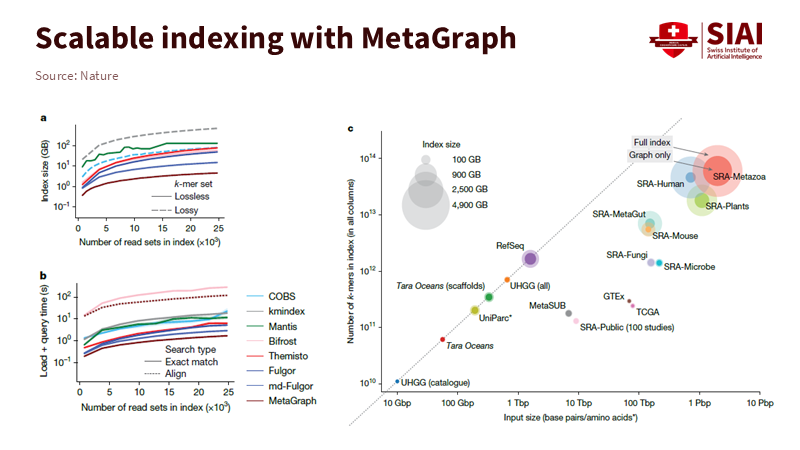
Note: As search scale increases, unit costs decline while accuracy remains stable
The Evolution of Search That Reveals Relationships
The focus of search technology is shifting from speed toward accuracy. MetaGraph maintains high recall even for sequences with high mutation rates or incomplete data. In human transcriptome analyses, it reconstructed positions and markers without requiring sequence realignment. This capability stems from evaluating the relational structure among datasets rather than isolated matches.
Graph-based search is evolving beyond result presentation into a method for analyzing relationships across data. In life sciences, it is used to link antibiotic resistance genes to phage hosts and to identify shared genetic signals and patterns within clinical datasets. Such approaches broaden interpretive scope and enable the discovery of new associations.
Graph-based search also establishes a new standard for data management. When the FAIR principles—Findable, Accessible, Interoperable, and Reusable—are embedded into the structure, the provenance and usage conditions of data can be traced with precision. When specific search paths consistently yield high accuracy or meaningful results, the system reinforces those paths as benchmarks for subsequent research. Ultimately, graph-based search is emerging as a technology that demonstrates how data is connected, rather than merely what is retrieved.
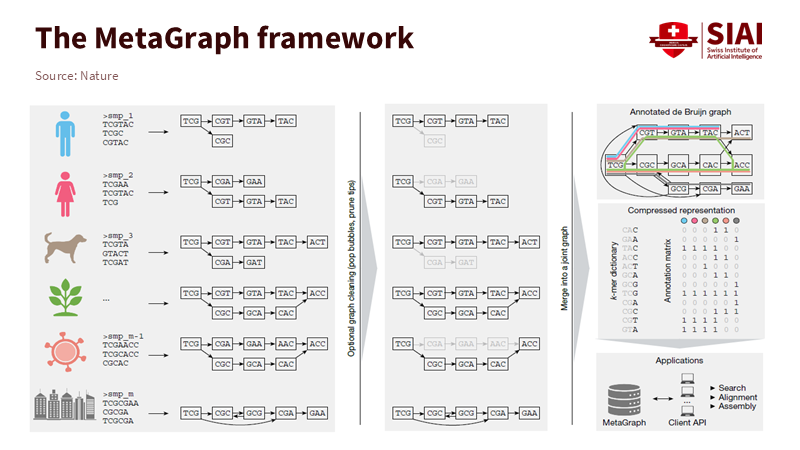
Note: Genomic sequences from different species are divided into k-mers, integrated into a connected graph, and stored in a compressed matrix format
Design and Trust Matter More Than Cost
The central challenge of data infrastructure is no longer cost. Graph-based search reuses redundant indices, improving efficiency as datasets expand. According to a report by the National Human Genome Research Institute (NHGRI), the cost of whole-genome sequencing has fallen below $1,000, with some cases approaching $500. This indicates that even university-level graph indices can be operated on standard servers or modest cloud environments.
The real challenge lies in design and trust frameworks. As technology advances, data governance systems must become increasingly sophisticated. Every node and path must clearly indicate provenance, usage conditions, and access rights, and indices require regular updates. During analysis or publication, a preserved “archival dataset” reflecting the data state at that time must be stored alongside results.
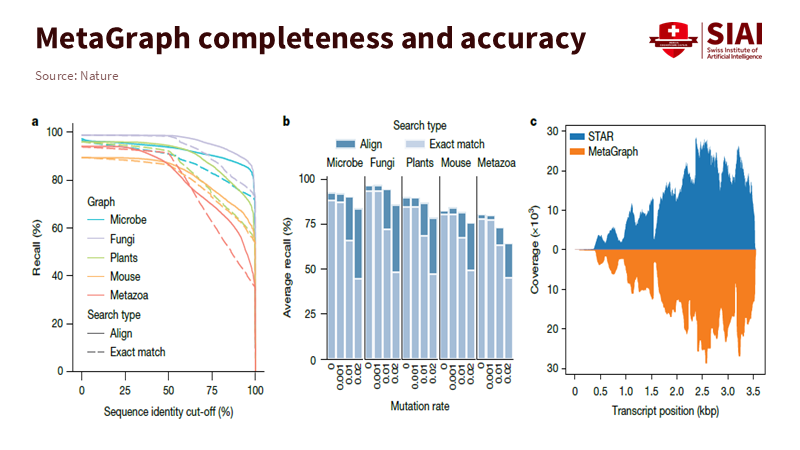
Note: MetaGraph maintains high search accuracy across all biological kingdoms, from microorganisms to animals, while ensuring uniform coverage across entire transcriptomes
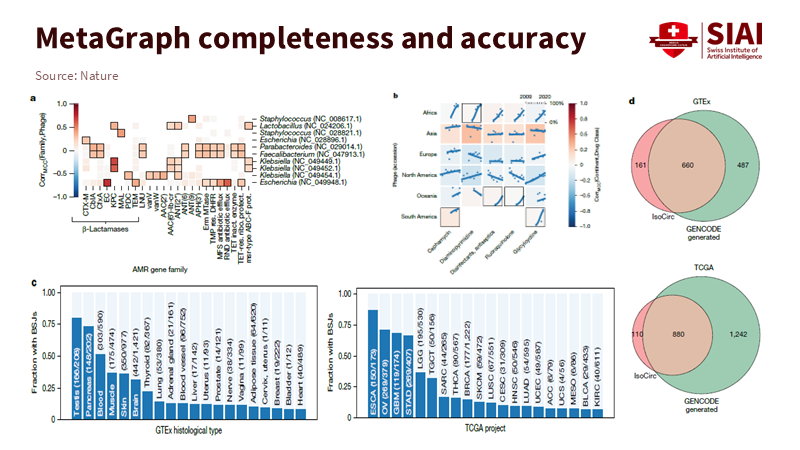
Note: MetaGraph precisely distinguishes antibiotic resistance genes and region-specific sequence patterns, demonstrating higher concordance than existing GENCODE references across GTEx and TCGA datasets
The Importance of Graph Literacy
As these technologies proliferate, graph literacy—the ability to understand how data is interconnected—becomes essential. Core concepts such as nodes (data points), edges (connections), paths (data flows), and colors (indicators of provenance or attributes) must be understood and applied in data structure design. With such literacy, graph-based data architectures can extend beyond life sciences into medical imaging, advanced materials synthesis, environmental sensing, and legal data.
Changing Structure Changes Discovery
MetaGraph has transformed biological information search by enabling petabase-scale exploration. Data now operates as graphs rather than lists, as networks of multiple interconnected paths rather than single matches. This shift transcends life science analytics, reshaping the fundamental principles of information processing and research methodology. Data scale and complexity no longer constitute limits. With the right structure, they become starting points for discovery. What is now required is systematic execution to extend this structural way of thinking into research and industry.
For the original version of this research article, please refer to DNA Search Engine for Learning, From Lists to Graphs. Copyright of this article belongs to the Swiss Artificial Intelligence Institute (SIAI).





















